A Biologist's guide to Bayesian phylogenetic analysis
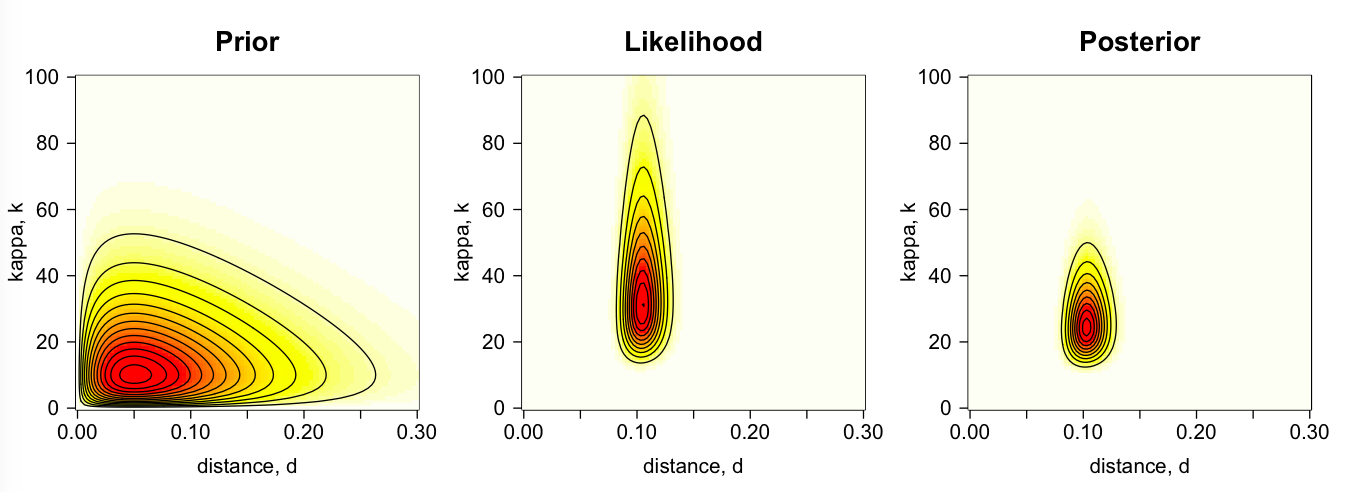
Our review paper on Bayesian phylogenetics is out! The review is intended for the biologist interested in using Bayesian phylogenetic tools to analyse their data. We cover topics such as specification of the prior, model selection, setting up the MCMC, diagnosing the MCMC chain (i.e. convergence, mixing, trace plots and the ESS), efficiency and autocorrelation of the MCMC, summarising the posterior and so on. We use a small example of estimating the molecular distance, d, and the transition-transversion ratio, k, in a two-species phylogenetic model, to illustrate the main concepts.
You can read the review for free here: rdcu.be/v4jM
A tutorial guiding through the two-species example is given here: bit.ly/BayesianNEE