BBSRC LiDO and NERC DTP PhD Studentship Projects
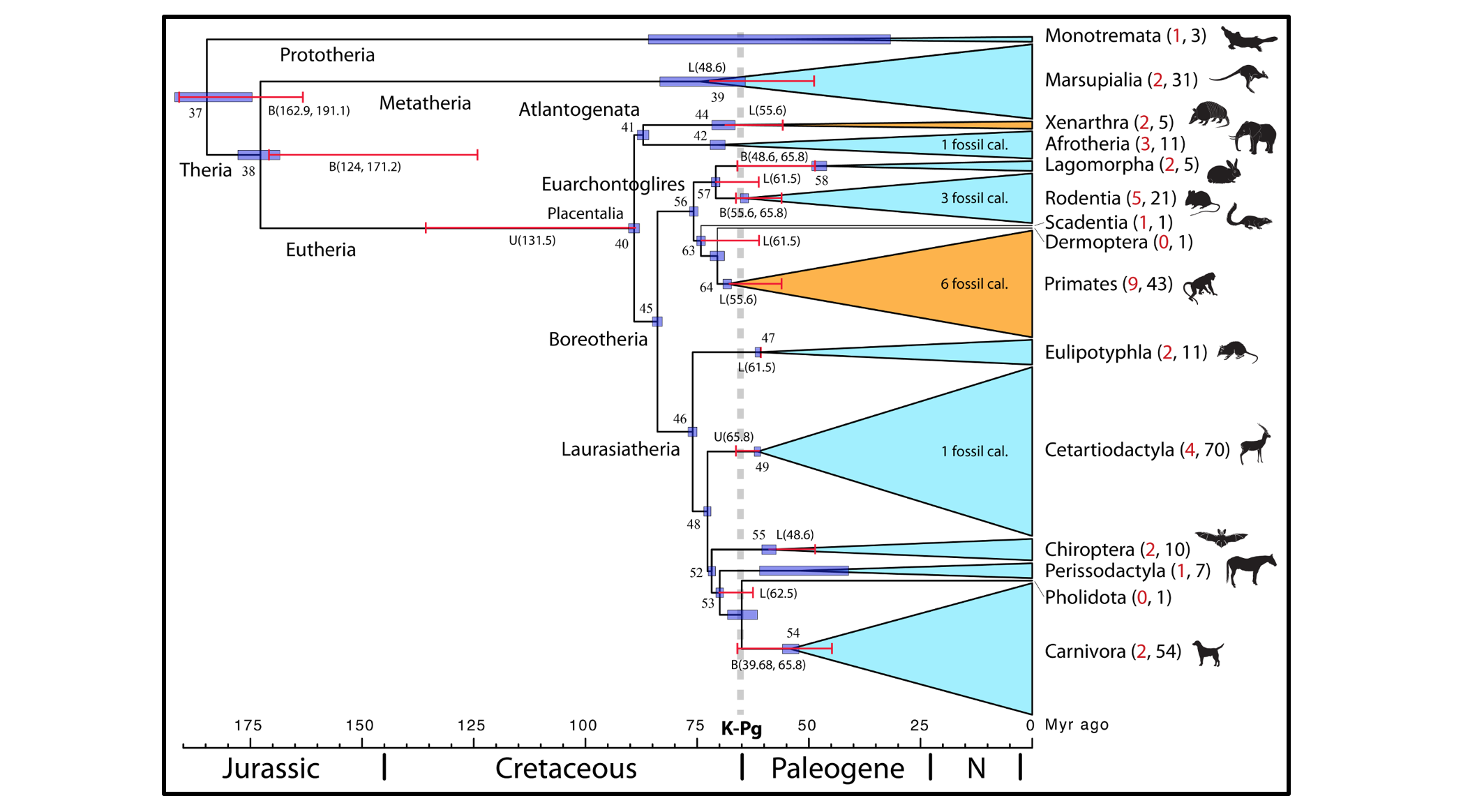
I currently have two PhD projects advertised in the BBSRC LiDO and NERC DTP programmes. Research in my lab focuses on the use of Bayesian statistics to model evolutionary processes. Right now we are focused on how to estimate species divergences by integrating fossils and genomes, modelling morphological evolution in phylogenies, and modelling the rate of evolution in phylogenies. If you are a student who has already been accepted for the 2018 LiDO or DTP starts, and you are interested in doing a project in my lab, please do not hesitate to contact me! Below you’ll find links to the two projects. The figure is a phylogeny of mammals, showing diversification timings relative to the great K-Pg extinction. The timings were obtained by analysing complete genomes (see our Proc. Roy. Soc. B paper below).
BBSRC LiDO: Bayesian statistical inference of divergence times
NERC DTP:Fossil morphology and genomic data to study species divergences through time
Key Publications
- dos Reis et al. (2016) Bayesian molecular clock dating of species divergences in the genomics era. Nature Reviews Genetics, 17: 71–80. DOI: 10.1038/nrg.2015.8
- dos Reis et al. (2015) Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Current Biology, 25: 2939–2950. DOI: 10.1016/j.cub.2015.09.066
- dos Reis et al. (2012) Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny Proceedings of the Royal Society B., 279: 3491–3500. DOI: 10.1098/rspb.2012.0683