Simple analysis
A nucleotide alignment of 16 species was simulated on the phylogeny of Figure 1A. The model is GTR+G with nucleotide frequencies T = 0.25318, C = 0.32894, A = 0.31196, G = 0.10592; alpha = 0.25; and substitution matrix parameters equal to 0.88892, 0.03190, 0.00001, 0.07102, 0.02418. Simulation software is Evolver v4.9e. Mean substitution rate is 1.
The data are then analysed with MCMCtree v4.9f to recover the divergence times under the JC69 and GTR+G substitution models using the approximate likelihood method and the strict clock. The rate prior is G(2, 2).
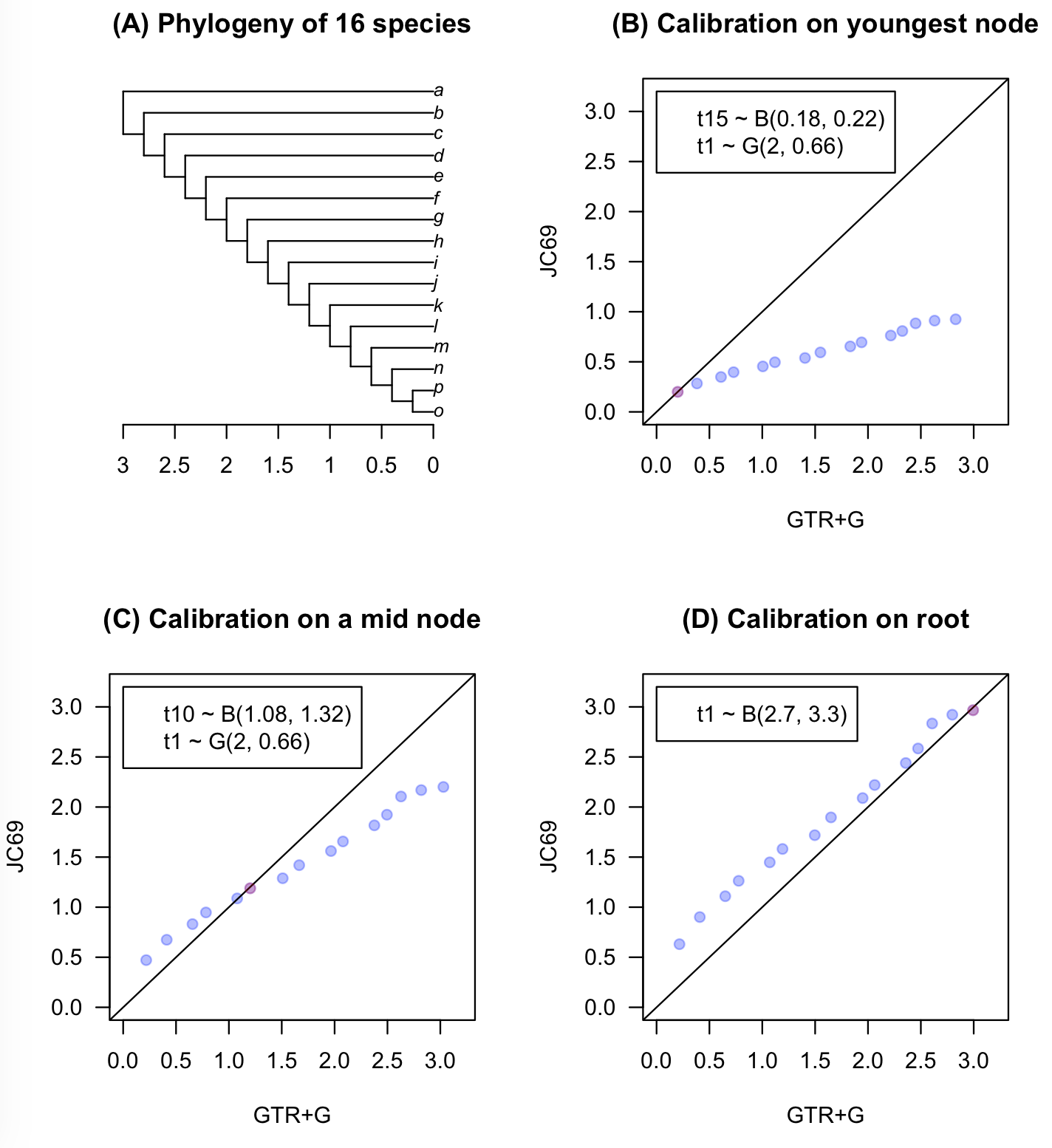
In Figure 1B, posterior mean times estimated under JC69 are shown in the y axis, while corresponding estimates for GTR+G are shown in the x axis. Times are estimated using a precise calibration on the youngest node in the phylogeny (red dot) and a very diffuse calibration on the root. The root calibration is G(2, 0.66) which has mean 3.03 (close to the true root age) and 95% prior CI (0.367, 8.44). In this case, there is catastrophic underestimation error when using the JC69 model (which is misspecified) in comparison to the GTR+G model (which is the correct model).
In Figure 1C, the precise calibration is moved to the sixth youngest node in the phylogeny (red dot). The effect of sliding the precise calibration to an older node is to “pull up” the estimated times under JC69 so that they are in better agreement with the estimates under GTR+G. However, there is still unacceptably large underestimation error for the ages of the oldest nodes.
In Figure 1D, the precise calibration has been moved to the root (red dot). Here both JC69 and GTR+G show broadly similar time estimates. In other words, a precise calibration on the root pins the inferred timespan of the phylogeny making the analysis robust to substitution model misspecification.
Figure 1:
Appendix:
Phylogeny used in the simulation:
(a:3.0,(b:2.8,(c:2.6,(d:2.4,(e:2.2,(f:2.0,(g:1.8,(h:1.6,(i:1.4,(j:1.2,(k:1.0,
(l:0.8,(m:0.6,(n:0.4,(o:0.2,p:0.2):0.2):0.2):0.2):0.2):0.2):0.2):0.2):0.2):0.2
):0.2):0.2):0.2):0.2):0.2);
Tree with fossil calibrations used:
# Informative calibration on youngest node:
16 1
(a,(b,(c,(d,(e,(f,(g,(h,(i,(j,(k,(l,(m,(n,(o,p)'B(.18,.22)'))))))))))))))'G(2,.66)';
# Informative calibration on mid node:
16 1
(a,(b,(c,(d,(e,(f,(g,(h,(i,(j,(k,(l,(m,(n,(o,p))))))'B(1.08,1.32)')))))))))'G(2,.66)';
# Informative calibration on root node:
16 1
(a,(b,(c,(d,(e,(f,(g,(h,(i,(j,(k,(l,(m,(n,(o,p)))))))))))))))'B(2.7,3.3)';
Time estimates (the true ages for the 15 nodes are 0.2, 0.4, 0.6, …, 2.6, 2.8, 3.0):
# Informative calibration on youngest node - JC69:
node t t025 t975 t025.hpd t975.hpd w.hpd
t_n17 0.9246 0.7374 1.1192 0.7326 1.1133 0.3808
t_n18 0.9112 0.7262 1.0998 0.7190 1.0911 0.3720
t_n19 0.8843 0.7046 1.0672 0.6988 1.0580 0.3593
t_n20 0.8074 0.6478 0.9707 0.6438 0.9651 0.3213
t_n21 0.7626 0.6131 0.9164 0.6113 0.9139 0.3026
t_n22 0.6945 0.5583 0.8349 0.5600 0.8360 0.2760
t_n23 0.6540 0.5263 0.7880 0.5180 0.7780 0.2600
t_n24 0.5939 0.4794 0.7154 0.4767 0.7121 0.2354
t_n25 0.5384 0.4346 0.6475 0.4312 0.6433 0.2121
t_n26 0.4956 0.4017 0.5947 0.3981 0.5898 0.1917
t_n27 0.4542 0.3678 0.5455 0.3635 0.5399 0.1764
t_n28 0.3966 0.3228 0.4750 0.3205 0.4722 0.1517
t_n29 0.3485 0.2862 0.4164 0.2840 0.4131 0.1291
t_n30 0.2838 0.2378 0.3339 0.2366 0.3321 0.0955
t_n31 0.1999 0.1800 0.2199 0.1802 0.2201 0.0399
# Informative calibration on youngest node - GTR+G:
node t t025 t975 t025.hpd t975.hpd w.hpd
t_n17 2.8292 2.0607 3.8926 1.9619 3.7189 1.7570
t_n18 2.6302 1.9624 3.4740 1.8940 3.3854 1.4914
t_n19 2.4503 1.8236 3.2318 1.7728 3.1654 1.3926
t_n20 2.3249 1.7360 3.0642 1.6732 2.9858 1.3126
t_n21 2.2156 1.6535 2.9121 1.6364 2.8797 1.2433
t_n22 1.9393 1.4553 2.5470 1.4271 2.5047 1.0775
t_n23 1.8326 1.3719 2.4133 1.3306 2.3465 1.0159
t_n24 1.5499 1.1505 2.0472 1.1294 2.0148 0.8854
t_n25 1.4043 1.0503 1.8555 1.0240 1.8149 0.7908
t_n26 1.1195 0.8351 1.4744 0.8106 1.4403 0.6297
t_n27 1.0053 0.7478 1.3266 0.7245 1.2942 0.5697
t_n28 0.7287 0.5416 0.9686 0.5345 0.9553 0.4208
t_n29 0.6095 0.4545 0.8047 0.4446 0.7891 0.3445
t_n30 0.3821 0.2916 0.4990 0.2832 0.4870 0.2038
t_n31 0.2002 0.1801 0.2201 0.1802 0.2202 0.0400
# Informative calibration on mid node - JC69:
node t t025 t975 t025.hpd t975.hpd w.hpd
t_n17 2.2005 1.9058 2.5448 1.9030 2.5383 0.6354
t_n18 2.1690 1.8807 2.5020 1.8807 2.5016 0.6209
t_n19 2.1052 1.8263 2.4340 1.8160 2.4215 0.6056
t_n20 1.9232 1.6746 2.2143 1.6583 2.1938 0.5356
t_n21 1.8170 1.5859 2.0877 1.5764 2.0750 0.4986
t_n22 1.6561 1.4507 1.8987 1.4424 1.8892 0.4469
t_n23 1.5606 1.3707 1.7804 1.3642 1.7725 0.4082
t_n24 1.4192 1.2565 1.6133 1.2524 1.6066 0.3541
t_n25 1.2886 1.1521 1.4507 1.1468 1.4439 0.2970
t_n26 1.1886 1.0786 1.3186 1.0765 1.3160 0.2395
t_n27 1.0875 0.9664 1.2239 0.9650 1.2218 0.2568
t_n28 0.9472 0.8299 1.0805 0.8266 1.0762 0.2495
t_n29 0.8309 0.7188 0.9578 0.7154 0.9538 0.2385
t_n30 0.6744 0.5715 0.7916 0.5701 0.7896 0.2195
t_n31 0.4712 0.3856 0.5690 0.3819 0.5644 0.1825
# Informative calibration on mid node - GTR+G:
node t t025 t975 t025.hpd t975.hpd w.hpd
t_n17 3.0291 2.4090 3.9177 2.3514 3.7879 1.4365
t_n18 2.8195 2.2762 3.4629 2.2428 3.4188 1.1760
t_n19 2.6283 2.1260 3.2244 2.0916 3.1796 1.0881
t_n20 2.4949 2.0247 3.0473 2.0036 3.0195 1.0159
t_n21 2.3760 1.9306 2.8986 1.9023 2.8591 0.9568
t_n22 2.0792 1.6959 2.5291 1.6605 2.4884 0.8278
t_n23 1.9657 1.6105 2.3806 1.5942 2.3595 0.7654
t_n24 1.6652 1.3659 2.0295 1.3541 2.0075 0.6534
t_n25 1.5091 1.2551 1.8247 1.2342 1.7938 0.5596
t_n26 1.2016 1.0800 1.3202 1.0806 1.3205 0.2399
t_n27 1.0785 0.8811 1.2604 0.8904 1.2670 0.3766
t_n28 0.7829 0.6289 0.9488 0.6218 0.9411 0.3192
t_n29 0.6557 0.5163 0.8084 0.5126 0.8027 0.2900
t_n30 0.4119 0.3196 0.5182 0.3156 0.5131 0.1974
t_n31 0.2180 0.1617 0.2835 0.1587 0.2788 0.1200
# Informative calibration on root node - JC69:
node t t025 t975 t025.hpd t975.hpd w.hpd
t_n17 2.9669 2.6969 3.2941 2.6910 3.2874 0.5964
t_n18 2.9218 2.6372 3.2606 2.6402 3.2632 0.6230
t_n19 2.8343 2.5355 3.1781 2.5458 3.1868 0.6410
t_n20 2.5846 2.2886 2.9241 2.2806 2.9150 0.6344
t_n21 2.4395 2.1520 2.7711 2.1385 2.7562 0.6177
t_n22 2.2199 1.9513 2.5277 1.9470 2.5220 0.5751
t_n23 2.0894 1.8282 2.3937 1.8197 2.3793 0.5596
t_n24 1.8965 1.6501 2.1767 1.6430 2.1673 0.5242
t_n25 1.7188 1.4927 1.9768 1.4820 1.9637 0.4817
t_n26 1.5811 1.3672 1.8228 1.3616 1.8166 0.4550
t_n27 1.4476 1.2446 1.6776 1.2358 1.6642 0.4284
t_n28 1.2629 1.0798 1.4716 1.0733 1.4633 0.3900
t_n29 1.1093 0.9440 1.2957 0.9433 1.2949 0.3516
t_n30 0.9010 0.7552 1.0664 0.7523 1.0628 0.3106
t_n31 0.6303 0.5145 0.7633 0.5096 0.7564 0.2467
# Informative calibration on root node - GTR+G:
node t t025 t975 t025.hpd t975.hpd w.hpd
t_n17 2.9943 2.6997 3.2979 2.6953 3.2928 0.5975
t_n18 2.7976 2.2899 3.2102 2.3657 3.2508 0.8851
t_n19 2.6069 2.1096 3.0547 2.1315 3.0686 0.9370
t_n20 2.4736 1.9939 2.9188 2.0132 2.9357 0.9225
t_n21 2.3575 1.8839 2.7978 1.9258 2.8326 0.9068
t_n22 2.0630 1.6377 2.4664 1.6697 2.4912 0.8215
t_n23 1.9494 1.5458 2.3333 1.5707 2.3547 0.7840
t_n24 1.6500 1.2951 2.0070 1.3040 2.0142 0.7102
t_n25 1.4964 1.1628 1.8283 1.1678 1.8312 0.6634
t_n26 1.1915 0.9212 1.4717 0.9197 1.4689 0.5492
t_n27 1.0707 0.8287 1.3210 0.8281 1.3202 0.4921
t_n28 0.7767 0.5950 0.9696 0.5868 0.9592 0.3724
t_n29 0.6499 0.4956 0.8132 0.4924 0.8096 0.3171
t_n30 0.4091 0.3067 0.5201 0.3049 0.5179 0.2130
t_n31 0.2166 0.1550 0.2852 0.1552 0.2852 0.1300